Deductive proof that self replicating systems inevitably evolve if they have a mechanism for passing on inheritable traits that allows for variation.
Introduction
My intention with this post is to lay out a deductive argument that shows that if you have a self replicating system of any type whatsoever, in which collections of states are passed along from one generation to the next via some mechanism of inheritance, and in which this mechanism for inheritance allows for variability within the collection of states that are passed down, then evolution is inevitable. I also intend to keep the main body of this argument as abstract as possible, so we won’t assume anything specific about the self replicating systems, or about what the states might be, or about what the mechanism for inheritance is or how or why it is variable. I’m going to try to keep the main body of my argument confined to the abstract worlds of math and computer programming. I will subsequently try to tie the argument to biology.
Definitions
Let’s begin with some definitions:
- Self replicating system (or SRS): a self replicating system is a system that is complete in and of itself that copies itself.
- Inheritability: the ability of an SRS to inherit its collection of states from the collection of states present within a parent of set of parent SRS’s.
- Variability: the tendency of the inherited collection of states not to be perfectly copied from one generation to the next.
- Tree style linked list: a set of nodes (which in this argument will represent SRS’s), which link to other nodes, in which parent nodes link to child nodes and child nodes link back to parent nodes.
- State Pool: A collection of states that is a union of all of the states contained in some collection of SRS’s
If there’s anything else you’d like me to define, or if anyone needs any of these definitions clarified feel free to leave a comment.
The Argument
So let’s say we have a self replicating system which, during the process of replication, copies some set of states that it contains to its child or copy. Let’s also say that the process of copying the set of states does not result in a perfect copy every time so that there is some variability in the states contained in the growing set of self replicating systems. Here we have a system with three main components. These components are self replication, inheritability and variability in the mechanism for inheritability.
One consequence that I will go into further a little later is that we start to have a growing state pool. I will only point out here that the state pool would remain constant and completely unchanging if there were no variability in the mechanism for inheritability when our self replicating systems replicated.
Another, which I’ll go into a little more now is that the resulting process will be a branching process which is analogous to a mathematical model known as a Markov Chain. Essentially a Markov Chain is a statistical model in which all of the information about the probability a system’s future states are contained in the system’s present state. This is analogous to the situation in our self replicating system in which the information in the the probability of what specific states will be present in the child is all present in the parent or parents. For the purposes of this argument, and because it serves as a “worst case” we’ll say that all possible single changes in the inheritable states of our SRS’s have the same probability, which effectively makes the variations random.
http://en.wikipedia.org/wiki/Markov_Chain
Markov chains introduce the terms transition, and transition probabilities. Transitions are the changes in state, in the case of our self replicating systems and their evolution the transitions would be the changes between parent and child which are present in the child. We could refer to the child as transitional, or containing transition states. The transition probabilities are the probabilities of the various transitions.
I believe it’s worth noting that even if the probability distributions for specific variations in states were not all equal, making the system not entirely random, it would not render the evolution of the system impossible, only nonrandom.
So to recap here:
We have a set of self replicating systems.
These systems pass along sets of states though a mechanism of inheritability to their copies (from parent to child).
The mechanism of inheritability contains some probability of mistakes during the copying process from parent to child.
This set of facts can be represented by a statistical model known as a Markov Chain in which the information about the probabilities of future states is contained within the present state.
The Markov chains generate a tree style linked list where parent node(s) link down to child node(s), child node(s) back to parent node(s), and, in the case of more than one parent, parent nodes link to each other. In this case where parent nodes link to each other their combination (or more precisely the union of the collection of states in all parents) establishes the “present state”.
The child nodes of our markov chain generated tree style linked list represent transitions each transition having, associated to it, it’s own transition probability.
If we don’t assume that all of our individual self replicating systems necessarily replicates or that of all of the self replicating systems which do replicate not all copy themselves the same number of times throughout their lifetime we can construct a diagram of a simplistic model of replication like this:
The above diagram shows a possible trend of replication starting with a single individual SRS. From this we can see that some individuals may be more successful than others at passing along its set of states from one generation to the next. We also see here that we have a growing state pool. Essentially the state pool can be considered to be growing as long as there are more nodes being added to the pool than there are being removed. Any aspect of any specific system which renders a SRS incapable of replicating effectively removes that SRS from the state pool. What may not be obvious from my diagram is that we should expect to see greater similarity between nodes that share a more recent common parent than we should from nodes that share a more distant common parent.
Here’s an example using a short string:
So in this example diagram we see that RABAGAJA and AABQGAJA both share a parent node and are much more similar to each other than they are to either of ATBPDQAA and ETBPAQAA which both share a parent node and are more similar to each other than they are to either of our first two strings. We can also see that ATBYLIAA is more similar to ATBPDQAA and ETBPAQAA than it is to either of RABAGAJA and AABQGAJA. This is due to the fact that ATBYLIAA share a more recent common parent node to ATBPDQAA and ETBPAQAA than it does to RABAGAJA and AABQGAJA.
It should be pretty easy to see that if the inheritable states in a set of SRS’s becomes more complicated, containing more variable information, then this situation will become more pronounced. This will be due to the fact that if the variation mechanism in the system is truly random then the probability that all of the same variations will eventually arise together independently in two distant branches gets smaller the more possible variations there are in the evolving system. This is because, if the mechanism for variability is truly random, the probability of any specific variation in the system is 1/P where P is the total number of possible variations. This describes an inverse relationship between the number of possible variations and the probability of any specific variation. So we see that as the number of possible variations increases the probability of any specific variation decreases.
I think it is also worth noting that this only necessarily holds true if the mechanism for variability is random. If the mechanism is nonrandom and some variations have a higher probability than others then it can become more likely that two distantly related branches will share these more probable sets of variations, and less likely that any will have or many will share the less likely sets of variations. To be clear here, this does not effect the evolve-ability of our systems here, it only effects the appearance of relationships between distant branches, because nonrandom variability introduces the possibility that distantly related branches can appear more closely related than they are. But this would have a noticeable effect on the distribution of the more probable variations in more of our evolving system’s isolated state pools. If some variations have a higher probability than others then we should expect to see these variations in a proportionally higher number of isolated state pools, even isolated state pools from distant branches in which other intermediary branches do not share the variation, or common parent nodes between the branches do not contain the variation.
Now back to what I was saying about the probability of specific variations. Here we have to keep in mind that the probability that from one step to the next there will be variation at all is going to depend on the nature of the variation mechanism, which we are not assuming anything specific about except that variations happen, which effectively eliminates a possibility of there being zero probability of variation. We can easily deduce that if the probability that will be variation from one step to the next is low then variations will be introduced slowly, possibly with several steps in a row having no variations between them at all. Conversely we can deduce that if the probability that there will be variation between steps is high then variations will be introduced more rapidly. It may also effect the transition probabilities if the probability that there will be variation was low but was calculated separately for each possible variation. In which case if the probability was higher than the number of total possible variations, for instance if the probability of a variation at each possible variation was 1/10,000 and the total number of possible variations was 100,000, then we might reasonably expect to see an average of 10 variations per step. On the other hand if the probability were lower than the total number of possible variations, for instance if the probability of a variation at each possible variation was 1/100,000 and there were only 10,000 possible variations we might reasonably expect to see an average of 1 variation every 10 steps.
And now if we focus on the probability of a variation happening when a variation does happen we can deduce a few more interesting things. For instance, if we look back at my diagram, we can see that once a variation has been introduced then as long as the branch in which it was introduced continues to be successful at replicating, and as long as the variation is not overshadowed by other variations, which becomes less likely the more possible variations there are in our SRS’s, then the variation will persist. As a result, though specific variations may have low probabilities of occurring, once they occur there is a higher probability that they may hang around a while and continue to be passed along throughout succeeding steps, unless the probability of replicative success is tied to certain aspects of an SRS’s states, in which case variations that negatively impact an SRS’s probability of replicative success will have a lower probability of being passed on to succeeding steps than variations that have no impact or have a positive impact.
Now if the probability that an individual SRS will replicate is random we can reach a few more interesting conclusions. One thing we can expect to see are changes in the distribution of states within the active state pool at any given step, with the SRS’s and branches that are more successful at replicating representing a proportionally larger share of the states in the active state pool (the active state pool being the state pool from which states are available to the next step). This can be seen in my last diagram as well. There you can see that the AACAAAAA branch was more successful at replicating than the ATAAAAAA branch, and we also see that there are more individual SRS’s with a C at the third spot than there are with a T at the second. In my last example diagram we also see that there is a variant from the AACAAAAA branch where the C state variant is replaced by a B state variant. This is an occurrence I will go into more later.
If we’re dealing with an evolving system in which there is a random chance that an individual SRS will replicate, and in which replication is coupled with a mechanism for inheritability, and in which variations are already present in the active state pool we’ll begin to see what I will call here state transition drift. State transition drift is a completely random process which cannot add state transitions to the state pool and will tend, instead, to remove them. Essentially the probabilities of any specific state variant becoming the dominant state within the state pool is proportional to its relative distribution within the active state pool. Thus in my last diagram the probability that the C state in the third position will become the only variant in the active state pool is 4/10, the probability that the B in the third position state variant will become the only state variant in the active state pool is 2/10 the probability of F in the third position becoming the only state variant in the active state pool is 1/10, and the probability of B in the third position becoming the only state variant in the active state pool is 3/10. The number of generations it might take for one state variant to become the only state variant in the active state pool is proportional to the probability that the state variant will become the only state variant in the active state pool and the number of SRS’s in the active state pool. This has to do with the law of large numbers. The effects of pure random chance on the outcome are also more pronounced in smaller populations because of the law of large numbers.
A simple example to illustrate this fact is a simple coin toss. Let’s say we toss a perfectly balanced coin 10 times. The probability of heads or tails is 1/2, let’s say we toss our coin 10 times and see these results: T, T, T, H, T, H, H, T, H, T. We have 6/10 tails and 4/10 heads, neither of which is 1/2. This is, in part, due to the fact that the probability of a heads or a tails result is the same each time I toss the coin. The same is the case for the replication of our SRS’s, the probability that an individual SRS will successfully replicate is the same for each SRS. The law of large numbers, specifically the strong law of large numbers, states that as the number of trials approaches infinity the outcome of the trials approaches the average. Since our populations of SRS’s will never reach infinity there will be some divergence from the averages that will inevitably result in some state variant losing ground to another until there is only one state variant, but if the populations are large this will occur more slowly, as the outcomes will be closer to the averages. In smaller populations there is a greater chance of divergence from the averages so state transition drift can play a more potent role in state variant distributions of smaller active state pools than larger ones. Once all state variations have been replaced by a single dominant state state transition drift cannot add new state variants to the active state pool, but remember for our discussion of state transition drift we left variability out. So once all state variants have been replaced by a single state in our active state pool new state variants can still be introduced by variability of our mechanism for inheritability. And it’s this capability of variability to add new state variants to the active state pool that I was drawing attention to when I pointed out earlier that the C state in the third position was changed to a B state in the third position in one of our steps in my last diagram.
Now let’s look at exchanges of state variants between active state pools. I will refer to this exchange of state variants as state variant flow. First let’s consider what happens if we have one active state pool that experiences a split wherein a subset of SRS’s that make up the active state pool are, for whatever reason, isolated from the active state pool. the result of this split is two separate active state pools. So we have separate active state pools, essentially, when we have more than one active state pool with no state variant flow between them. We can, due to state transition drift and variability, expect these separate active state pools to diverge from each other. This is because replication is random (in our worst case set of assumptions), as is variation in the inherited states (again according to our worst case assumptions). The smaller the break away active state pool is the greater the impact that state transition drift will have, for the reasons already discussed, and variability being random means that the probability of a given variation being mirrored in all or even most of the separate active state pools after they have broken away from one another decreases as the number of possible variations increases. The result of these two facts will be divergence between the separate active state pools over several steps.
Now if two active state pools that have been separated for several steps are brought back together then, we may expect to see one of at least two things happen, not assuming anything specific about our SRS’s or the mechanism for inheritability. Either they will no longer be compatible for replication, in which case the two active state pools will remain isolated even if they were to meet again, since neither could contribute state variants to the other. Or state variant flow would continue between the two active state pools and we should expect to see the action of state transition drift begin working to average out the differences between the two active state pools. We should also expect the effect of state variant flow between the sets of active state pools to be proportional to the proportion of one of both of the active state pools that are mixing. Thus it should have a much slower and less pronounced effect if there is only a small amount of state variant flow between the active state pools, and more pronounced and rapid if there is a large amount of state variant flow between the active state pools, notwithstanding the role of the law of large numbers on the rate of change in state transition drift. From all of this we can deduce that as state variant flow increases between active state pools the probability of divergence decreases between active state pools.
So let’s recap again briefly before we move on to selection:
We have self replicating systems.
These self replicating systems have a mechanism by which they pass on inheritable traits to their copies.
The mechanism for passing on inheritable traits has the capacity to copy those traits imperfectly introducing variations in the inheritable states being passed down.
This system of self replication wherein all information about the traits to be inherited with some probability of state variation by the next step in the evolving system is completely contained in the current state of the evolving system satisfies the Markov property, so our evolving system is a Markov chain.
Markov chains generate tree style linked lists as defined earlier and described in my first diagram.
We are assuming as a worst case assumption that variability is random.
We were, until now, since we had not introduced the concept of selection, assuming that replication was random.
State transition drift will, if we assume that replication is random, tend to remove state variants randomly from the active state pool (again the active state pool being the state pool that is capable at this step of contributing states to the next step).
State variant flow is the exchange of state variants between active state pools, it will tend to work against those active state pools significantly diverging from one another.
So now that we are all, hopefully, on the same page, let’s introduce the concept of selection to our evolving system. Let’s simply define selection as a weighting of the probabilities either for or against an individual SRS’s replication. We can further tie this weighting to the inheritable states of our SRS’s in order to make it relevant to the system’s evolve-ability. The reasoning here being that if the weighting of the probability for or against the SRS’s replicative success is not inheritable then it’s either random or arbitrary and doesn’t add anything to our understanding of the evolving system. An example of a selection event that weights an individual SRS’s probability of replicative success but is not inheritable would be the accidental death of a competitor with a higher probability of replicative success. The loss of that competitor may increase the remaining individual’s probability of replicative success, but that kind of chance event can’t be inherited.
Selection pressures will tend to be nonrandom consistently effecting many steps on our Markov chain. They may also tend to change slowly or oscillate between a set of states according to some set of rules. There may also be multiple selective pressures which may effect different SRS’s more than others and in different ways. SRS’s may even compose the majority of the selective pressures for other SRS’s (as in the predator/prey relationship). In systems where inheritable states have to meet some minimal fitness requirements for an SRS to be viable (or able to perform minimal functionality), one selective pressure may exist in its own inheritable states, there being a possibility for certain variations of those states to render the individual SRS nonviable, or at the very least give it a 0 or at best low probability of replicative success.
Now if we look again at state transition drift without the assumption that replication is random we see that it turns into an optimizing function with selective pressures weighting the probabilities of individuals SRS’s with certain state variants either favorably or unfavorably. This leads to an increased probability of state variants that contribute to an increased probability of replicative success replacing those state variants which provide no net improvement over the status quo or a decrease in the probability of replicative success. Again the law of large numbers will still apply, so the changes will be more gradual in larger populations unless the increase in probability of replicative success is drastic enough to significantly alter the odds. And I should also add that with selection as a consideration the trends of state transition drift may tend to change as selective pressures change. This is due to the fact that as selective pressures change the state variants that provide an advantage versus those which provide a disadvantage may also change.
Conclusion
Essentially evolution is an optimizing process which is made possible and inevitable by self replication, a mechanism for the inheritability of sets of states, where that mechanism is prone to variability. Especially where the states serve as, sort of, “solutions” to some type of “problem”. As in the case of organisms (for example), a “problem” would have been ion transport through lipid bilayers, leading to the “solution” of the evolution of proteins which bind to the lipid bilayer and act as ion channels. This “solution” would have been, by no means, intentional, rather the product of random changes in the genome of the first cells, but one which, once existent, would have drastically improved the probability of replicative success for the individual with this gene. This would lead to weighting in the state transition drift process that would see this trait become dominant throughout the resulting active state pool after several steps down the resulting Markov chain. These links are to concepts that were too specific for this blog entry, but that my abstract descriptions, nonetheless, fit. I didn’t say anything specific at all about mutation because that would have required that I assume a lot of very specific things about the mechanism by which variations happen in my self replicating systems. Speciation could be seen as the case, in my state variant flow example, in which when two isolated active state pools come back into contact with one another but are no longer compatible, thus remain isolated despite being in contact again.
http://en.wikipedia.org/wiki/Genetic_drift
http://en.wikipedia.org/wiki/Gene_flow
http://en.wikipedia.org/wiki/Mutation
http://en.wikipedia.org/wiki/Natural_selection
http://en.wikipedia.org/wiki/Speciation
If anyone has any criticisms, questions or suggestions feel free to comment.
EDIT: Someone suggested that I add another recap to sort of tie this post up into a nice little bow. And that’s actually a good idea. So a final recap of the entire post follows:
If we have a self replicating system, and that self replicating system has a mechanism for inheritability and that mechanism for inheritability is subject to imperfect copying which introduces variability;
And if the inherited traits come directly from a copy’s parent or set of parents;
Then our self replicating systems satisfy the Markov Property.
And if our self replicating system’s satisfy the Markov Property, then we can form Markov chains with them.
If we can form Markov Chains with our self replicating systems and if we assume (for the sake of using a worst case scenario) that variability is a random process (that all possible variations have an equal probability of happening), then we should expect variations to be randomly added into our self replicating system’s active state pool.
And there is a form of selection. This form of selection will tend to be constant over spans of time, or it may oscillate relatively slowly, or it may change according to a somewhat regular pattern, essentially it will tend not to be random or completely unstable. There may also be many selection pressures which may or may not overlap each other entirely or in part in some set of domains.
And if this form of selection acts to modify the probabilities that individual self replicating systems will successfully replicate, and if the states which the selection pressures are basing their selection on are inheritable states then we will see self replicating systems which have states which favorably weight the probabilities that a self replicating system will replicate tending to replicate more often, and we will see an increase in the relative distribution of self replicating systems with the favorable trait in the active state pool.
And we will also see self replicating systems with states which unfavorably weight the probabilities that a self replicating system will replicate tending to replicate less often. As a result we will see a decrease in the relative distribution of self replicating systems with the unfavorable trait in the active state pool.
Self replicating systems may move to selection domains where they are already suitable or random state variants may come into the state pool through variability which improve the self replicating system’s suitability for the selection domain it finds itself in.
If there is migration to another selection domain by some self replicating systems which comprise an active state pool but not all, and if the migration renders the two resultant active state pools replicatively isolated, resulting in no state variant flow between them, then we will expect to see divergence.
This will be because variability is random and so has a low probability of being the same in both active state pools, with the probability decreasing as the size of the set of inheritable states increases.
If one of these active state pools is small then state transition drift will have a larger role to play in determining what set of state variants becomes dominant within that active state pools, with state transition drift playing a larger deciding role the smaller the active state pool, though not without some consideration for the impact of selection. Essentially states with less of a major impact on replicative success even with selection will be the states between which state transition drift will ultimately be the deciding factor.
Over several steps along the Markov chain of these two separate active state pools new state variants will have been introduced to both active state pools through random variation, differing selection domains will result in different sets of state variants being optimal in each active state pool, and the random nature of state transition drift on those state variants that have less of an impact on replicative success even with selection will be found in both active state pools in different degrees of relative distributions. We might say the two active state pools are diverging.
If the two active state pools were to come back into contact with each other, and if they were still capable of exchanging state variants between each other then with the resumption of state variant flow between the two active state pools we will see the effect of state transition drift working to average out the differences between the two active state pools, effectively reducing the amount of divergence between them.
We will also see the rate at which state transition drift works to average out the differences is proportional to the degree of state variant flow between the two active state pools. So if there is a higher degree of state variant flow between the two active state pools then state transition drift will result in faster averaging of the differences between them, and if there is a smaller degree of state variant flow then there will be slower averaging of differences.
Therefore we can see that the net result of a self replicating system, which contains sets of traits which are inheritable by its copies in which the mechanism for inheritability is subject to imperfect copying resulting in variability within the inherited sets of states when coupled with selection results in an optimizing function which will tend to see beneficial state variants represented in a relatively higher proportion of the self replicating systems which make up an active state pool than non-beneficial state variants. This optimization function is analogous to biological evolution.
Explore posts in the same categories: Biology, Logic, Science
Tags: argument for evolution, Biology, Evolution, Logic, logic of evolution, proof of evolution
You can comment below, or link to this permanent URL from your own site.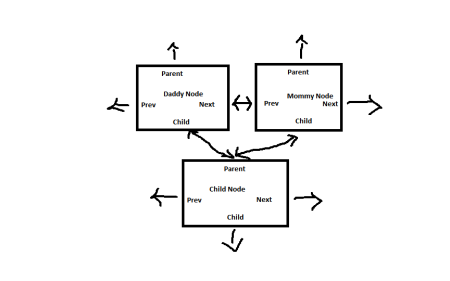
October 30, 2010 at 9:28 pm
[…] As for why cosmology shouldn’t be considered part of evolution…evolution is a process which requires 3 things. 1) self replicating systems, 2) the self replicating systems must have traits which can be inherited by the next generation of the system by some mechanism, and 3) this mechanism for inheritability of traits must be variable by some mechanism. Then if the system satisfies the Markov property (as our biological systems do) evolution becomes an inevitable result. I wrote an argument for evolution being inevitable in any system which meets these requirements here: https://intelsblog.wordpress.com/2009/12/24/proof-of-evolution/ […]
October 30, 2010 at 9:32 pm
If I were to have waited till today to write this I would have included satisfaction of the Markov property as a requirement for evolution. The overall argument would still run the same, however.